FALL 2021, THE EVIDENCE FORUM, WHITE PAPER
 Lindsey Wilson, BS PPAS Data Collection, Real-World Evidence Evidera, a PPD business |  Payal Pozin, MPH Research Associate III, Real-World Evidence Evidera, a PPD business |  Margaret Richards, PhD, MPH Senior Research Leader Real-World Evidence Evidera, a PPD business |  Ben Greener, MS, PharmD Senior Director Clinical Science Strategic Development Consulting PPD |
Selective Pressure Drives the Emergence of New SARS-CoV-2 Variants
Viruses are infectious biological agents that, in their most basic form, consist of a protein coat that encapsulates genetic information as either DNA or RNA. Once a virus enters a host, its primary function is to replicate while escaping clearance by immune mediated mechanisms. Effective replication may lead to more transmissible particles being shed, greater spread to other hosts, and continued survival. Given that viruses tend to travel light due to their limited size, they must rely at least partly on host machinery for their replication and spread. Viruses therefore hijack host cellular machinery to assist in making copies of their genetic information and producing the proteins that their genome encodes. The more a virus circulates in a population, the more likely it is that errors will be made while replicating its genome, resulting in asting changes to its genetic material – otherwise known as mutations. These “errors” can be due to the natural error rate from polymerases in the host or may be mediated by specific viral mechanisms that accelerate this process, depending on the virus.1 As the cycle of replication is short for most viruses, these mutations can occur rapidly due to the sheer volume of progeny being produced. Further, these mutations continue to accumulate over time as more replication cycles occur for a genome that has already been altered from its original sequence. Changes in the genetic code of a virus can result in structural changes to the encoded viral proteins that may alter the set of observable characteristics of the virus, such as transmissibility, clinical presentation or progression, efficiency of replication, or pathogenicity. Sometimes, mutations emerge with no impact on the viral protein sequence; these are regarded as silent mutations. Other times, changes in the amino acid sequence alter how the virus behaves and may give the virus either an evolutionary boost over its siblings or alternatively, may be detrimental to replication and survival. These accumulated mutations can confer an increase or decrease in the overall fitness (a comparative measure of survival advantage) of that virus and may lead to dominance for viruses with a substantial advantage. Throughout the SARS-CoV-2 pandemic, we have already observed that this evolutionary boost may result in increased transmission, evasion of natural immune response or neutralizing antibodies, and other phenotypes that impact public health and social policy requirements.2 The key to reducing viral mutations is simple: limit the host reservoir and therefore viral proliferation. This is achieved by decreasing transmission through policy and reducing the potential for effective replication through vaccination.
SARS-CoV-2, the virus that causes COVID-19, has been circulating worldwide since early 2020. Since the start of the pandemic, scientists and public health officials have been actively monitoring the evolution of the SARS-CoV-2 virus through genomic surveillance (the process of decoding the genomic sequence of the virus in clinical isolates and observing how it changes over time). Genomic surveillance helps public health officials track the path of the pandemic and identify mutations of concern. SARSCoV-2, during its widespread transmission and replication throughout the pandemic, has gone through countless replication cycles resulting in many accumulated changes, which are continuously subjected to a range of selective forces that allow for new variants to emerge and gain dominance. This process, Darwinian evolution, is caused by natural selection acting upon variations of mutations, resulting in the selection of mutations that make the virus more fit to compete, survive, and reproduce. For viruses, this selection process is well characterized and can be replicated in a laboratory by various approaches including the application of selective pressure in experimentally infected cell cultures.1
Figure 1. Genetic Drift and Selective Forces During Viral Replication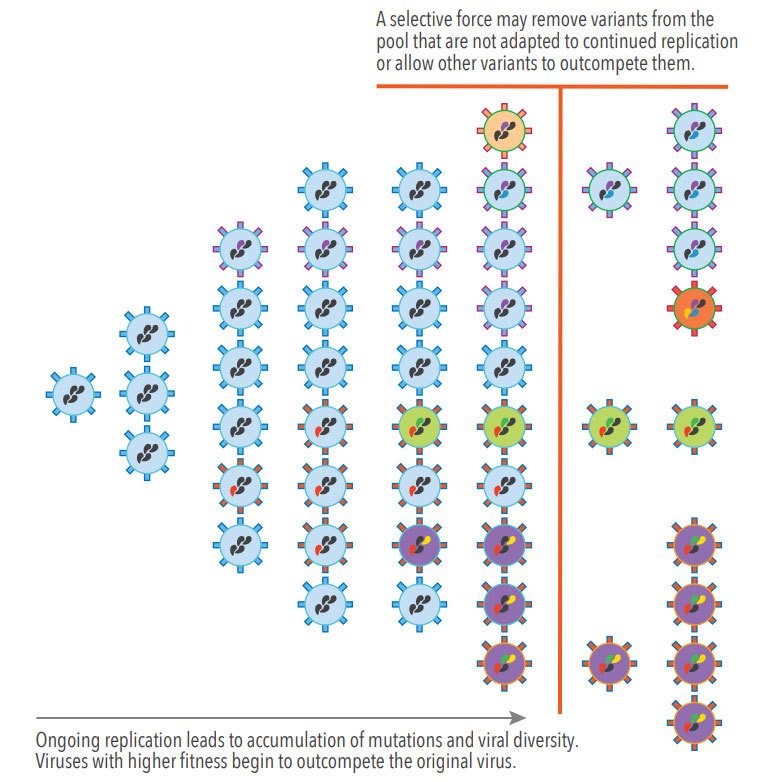
In the early stage of the COVID-19 pandemic (late January to early February 2020), scientists identified the D614G mutation, a problematic change to the genomic sequence at the 614th amino acid position of the spike protein, where the amino acid aspartate (D) was supplanted by glycine (G).3 This change to the spike protein resulted in enhanced viral loads in the upper respiratory tract that may have increased viral shedding and therefore transmissibility.4-6 When mutations such as D614G make a virus more transmissible, the mutated virus will continue to proliferate and eventually become more prevalent among the population at large, competing with other variants of the virus and crowding out less transmissible variants. As of June 2020, the D614G mutation was identified in nearly all SARS-CoV-2 samples worldwide and was present in dominant circulating viruses along with other mutations.7 It is important to note that while a single mutation can certainly generate a new phenotype, the stepwise process of accumulating mutations and the sum impact of these changes can profoundly change the behavior of a pathogen. We can group sets of these changes into viral clades and lineages. These biological terms are ways to classify groupings and genetic relationship. A clade is a genetic grouping that can be traced back to a single ancestor while a lineage refers to a continuous line of descent within a clade. Grouping mutated virus sequences by lineage is helpful at the front edge of the pandemic, during outbreak investigations, or when tracking patterns on a more granular scale. Lineage classification takes external information into account, such as details on how the virus is spreading and the genetic sequence data. What we now refer to as the “Alpha Variant” or “Delta Variant” are actually described by lineages – specifically, lineage B.1.1.7 and B.1.617.2.
In addition to increased transmissibility, there are a number of reasons why specific variants become increasingly dominant in the viral pool. Some combinations of mutations may allow the virus to escape the activity of neutralizing antibodies or drugs due to a structure change in a viral epitope or decreased affinity at the target site for small molecule therapeutics. In some cases, specific mutations may assist viruses in overcoming natural immunity or therapeutic or prophylactic interventions. This is because only viruses that can escape immune detection or avoid neutralization are able to replicate effectively so a selective pressure is introduced. Such a scenario is a risk among vaccinated individuals, who may inadvertently be amplifying more virulent pathogens or resistant pathogens by suppressing proliferation of competing viruses that are more susceptible. It is important to understand that while this type of selection is possible in vaccinated individuals, the burden of mutation created in individuals who are not vaccinated is far higher since replication may occur at higher levels. The process of viral evolution is the reason why it is so important for an entire population to get vaccinated, as opposed to only a portion of the population, in order to decrease infections worldwide and slow the emergence of new variants that may be able to overcome the barrier of vaccination and therapeutics. While it is expected that new variants of SARS-CoV-2 that pose an increasing risk to humans will continue to emerge, continued genomic surveillance and prediction tools will help mitigate this risk.
Download White Paper
Fill out the form below to download this whitepaper.